Post Electrophoretic Analysis Articles
Differential Display
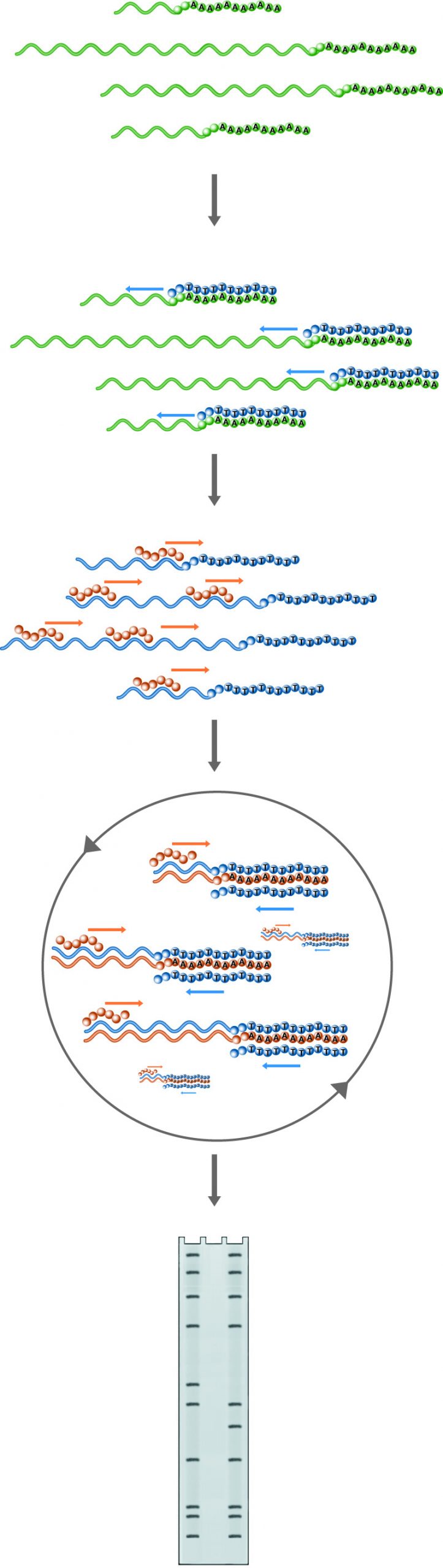
The differential display is a PCR-based technique that generates a characteristic set of DNA fragments from the messenger RNA pool within a cell. The use of random hexamers (brown) in combination with oligo-dT primers (blue) allows the amplification of a population of DNA species which changes with the composition of the starting RNA pool. The differential display is a powerful tool for the analysis of gene expression.
Differential display is a powerful technique for detecting and quantitating changes in gene expression patterns between differently treated cells. Fragments of those genes which are induced or suppressed can be identified and isolated for further analysis, with no prior knowledge of the sequences involved. The technique is PCR based, and yields results in only 1 - 3 days.
The differential display is a variation of standard PCR, allowing the amplification of a large population of fragments, rather than the specific amplification of one band. Specificity is reduced in two ways. First, random primers are used at one end of the amplification. A mixture of all possible hexamers (46 =4096 primers) is used. This allows many different molecules to be amplified.
The second drop in specificity is provided by using the poly-A tail present on the vast majority of RNA species. A poly dT primer is used to prime on this tail. The 3' end of the poly dT primer has a pair of non-T bases, which is sufficient to "anchor" the primer to the end of the coding sequence. There are thus 32 = 9 different possible primers at this end.
Amplifications run with random hexamers and the poly dT primers on cDNA (that is, DNA copied from mRNA) taken from treated and untreated cells, give a pattern of bands, each of which represents an amplified fragment of an expressed gene. A fragment that appears in one sample but not the other is either induced or suppressed by the treatment. Such bands may be cut out and re-amplified (using the same primer set as in the original reaction) for further analysis including sequencing, cloning, probe synthesis, etc.
The differential display is covered by patents owned by GeneHunter. GeneHunter recommends the use of SequaGel - UreaGel 6 for differential display analysis.
NEXT TOPIC: RNA Mapping
- Using PAGE to Determine Nucleic Acid Molecular Weight
- SSCP Analysis
- Sanger Sequencing
- Sample Preparation for Native PAGE of DNA
- Sample Prep for Denaturing PAGE of DNA
- S1 Mapping
- Run Conditions in Denaturing PAGE
- RNA Mapping
- RNA Electrophoresis
- Ribonuclease Protection
- Restriction Digest Mapping
- Primer Extension
- Preparing Denaturing DNA & RNA Gels
- Preparation of Denaturing Agarose Gels
- Preparation of Agarose Gels
- Pouring Sequencing Gels
- PFGE and FIGE
- PCR Analysis: Yield and Kinetics
- PCR Analysis: An Examination
- Native PAGE of DNA
- Mobility Shift Assay
- Methylation & Uracil Interference Assays
- Maxam & Gilbert Sequencing
- Manual Sequencing
- In Gel Enzyme Reactions
- Heteroduplex Analysis
- Gel Preparation for Native PAGE of DNA
- Gel Electrophoresis of PCR Products
- DNase I Footprinting
- DNA/RNA Purification from PAGE Gels
- DNA/RNA Purification from Agarose Gels – Electroelution
- Differential Display
- Denaturing Polyacrylamide Gel Electrophoresis of DNA & RNA
- Conformational Analysis
- Automated Sequencers
- Analysis of DNA/Protein Interactions
- Agarose Gel Electrophoresis of DNA and RNA – Uses and Variations
- Agarose Gel Electrophoresis of DNA and RNA – An Introduction
