Post Electrophoretic Analysis Articles
DNA/RNA Purification from PAGE Gels
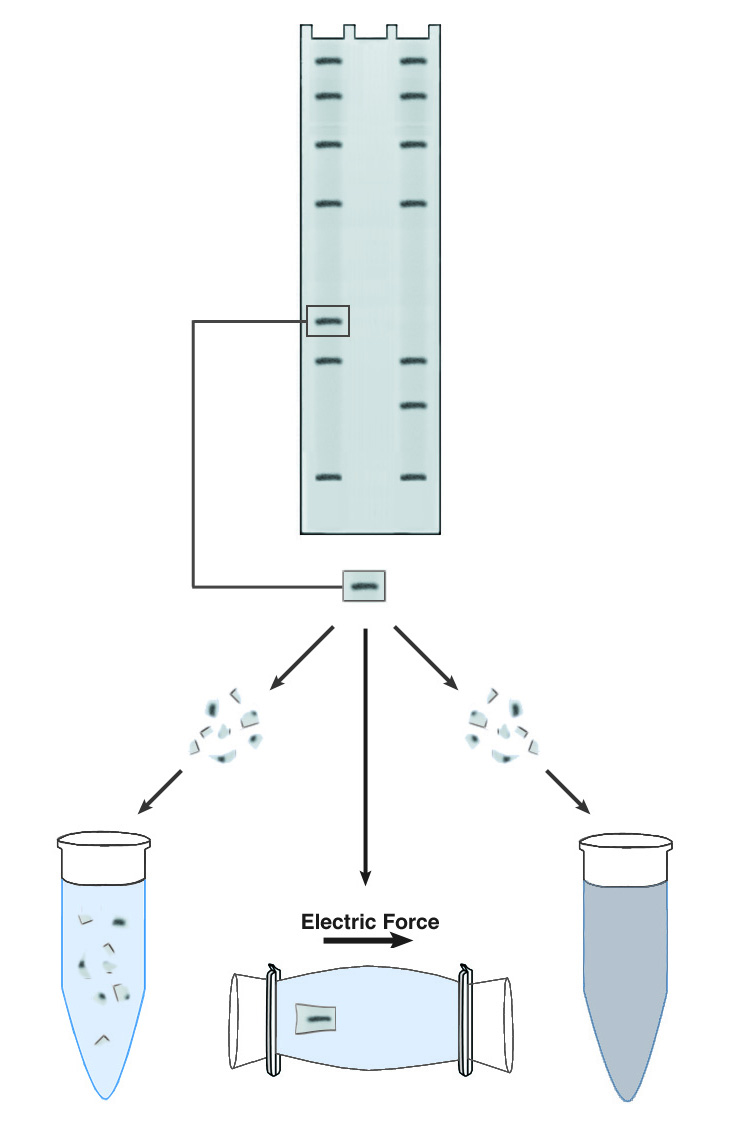
Methods of DNA/RNA Purification from PAGE Gels.
Polyacrylamide gel electrophoresis yields individual bands of extremely high purity, in that only one nucleic acid sequence tends to be present in each. Electrophoresis can thus serve as a powerful purification tool for DNA or RNA. In practice, most purifications are carried out on DNA, which is more stable and tends to be available in larger quantities. A major obstacle to the purification of DNA using PAGE is the recovery of the DNA after electrophoresis. By definition the sieving polyacrylamide matrix used in PAGE offers significant resistance to the free diffusion of DNA molecules, and vigorous steps are required to separate the DNA from the matrix.
The first step in all recovery protocols is to cut out the band of interest mechanically. The band is visualized, usually with ethidium bromide, and then excised from the gel with a razor blade or a glass coverslip. It is important to trim the band of all excess gel material, because the purity and dilution of the recovered DNA are greatly affected by the volume of the initial gel slice. The slice may be soaked briefly in water to remove gel buffer and ethidium bromide, prior to recovery of the DNA.
The simplest approach to recovering DNA from PAGE gels is known descriptively as "crush & soak." This technique accelerates the slow diffusion of DNA from a block of polyacrylamide by maximizing the surface area for contact between the gel and the elution buffer. The gel slice is crushed into a fine slurry, which is soaked in the elution buffer of choice. The technique is easy to use and requires no special equipment, but recovery is usually low (30-50%) and requires long incubations, generally 18-36 hours. One significant advantage is that the elution buffer does not need to have any special characteristics. The DNA can be recovered in water if desired.
Another commonly used technique is electroelution. This is effectively a second electrophoresis step, which causes the band to migrate out of the matrix and into the surrounding buffer. The DNA recovered by electroelution is generally of very high purity, and recoveries can approach 90%. The technique is also rapid, as the DNA need only migrate 2-3 mm to be freed from the gel. Electroelution requires specialized apparatus or relatively complex gel manipulations, and the elution buffer must have a high enough conductivity to support electrophoresis, so the DNA recovered requires further purification to remove salts. Depending upon the apparatus used, the DNA may be recovered into a relatively large volume. Ethanol precipitation is often used to concentrate the DNA and remove buffer salts.
Crush and Soak
The excised, trimmed gel slice is crushed or cut to produce pieces less that 1mm in any dimension. Alternately, the gel slice can be forced through a small opening, ie through a syringe needle. A convenient method is to use a hot needle to make a hole in the bottom of a 0.5ml microcentrifuge tube. The gel slice is placed in this tube, which is placed in turn into a 1.5ml tube. The assembly is centrifuged at top speed (14k rpm) for 1-5 minutes, or until the entire gel slice is collected in the lower tube.
Sufficient buffer is added to the gel pieces to cover them, and this slurry is incubated with shaking for 2-12 hours. Longer incubations give better recoveries, and are essential for acceptable recovery of fragments larger than 500bp. The slurry is then centrifuged and the supernatant recovered with a pipette. Polyacrylamide fragments carried over into the supernatant may be recovered by a second centrifugation step, or by filtration through a 0.2mm filter.
Electroelution
Various apparatus are available to make electroelution faster and easier, as well as to promote recovery of the DNA into as small a volume as possible. A method is presented below for performing electroelution in a standard horizontal gel box. It is based upon the fact that DNA molecules cannot pass through a dialysis membrane, while buffer components can. The DNA slice is placed in a bag of dialysis membrane containing a minimal amount of buffer. A voltage is applied, and the DNA migrates out of the gel and is trapped and recovered inside the dialysis bag.
- Prepare 0.3 liter of 0.1X TBE
- Place the gel slice in a small bag of dialysis tubing. The bag may be knotted to form the ends, but using dialysis clips is easier and the clips are convenient for handling the bag.
- Add a minimal volume of 0.1X TBE Use just enough to surround the gel slice. Seal the bag, taking care to exclude all air bubbles.
- Fill the horizontal mini-gel apparatus with 0.1X TBE, and place the dialysis bag in the apparatus, on the gel platform. Place the bag so the DNA slice is closest to the negative (black) electrode. Note: some dialysis clips float - it may be necessary to weigh the bag down with glass slides. Do not use metal as a weight, because it distorts the electrical field. Alternatively, the bag may be taped (on the clips!) to the gel platform to keep it from floating.
- Apply 2V/cm to the apparatus, and allow the DNA to elute at this potential for 2-3 hours.
- Recover the bag, open carefully and recover the buffer (containing the DNA) with a pipette. Rinsing the bag and gel slice once with 200 µl of 0.1X TBE will increase recovery substantially.
- If desired, add 0.1 vol 3M sodium acetate, and 3 volumes ethanol. Pellet the DNA and wash one time with 70% ethanol. Allow to air dry.
NEXT TOPIC: Conformational Analysis
- Using PAGE to Determine Nucleic Acid Molecular Weight
- SSCP Analysis
- Sanger Sequencing
- Sample Preparation for Native PAGE of DNA
- Sample Prep for Denaturing PAGE of DNA
- S1 Mapping
- Run Conditions in Denaturing PAGE
- RNA Mapping
- RNA Electrophoresis
- Ribonuclease Protection
- Restriction Digest Mapping
- Primer Extension
- Preparing Denaturing DNA & RNA Gels
- Preparation of Denaturing Agarose Gels
- Preparation of Agarose Gels
- Pouring Sequencing Gels
- PFGE and FIGE
- PCR Analysis: Yield and Kinetics
- PCR Analysis: An Examination
- Native PAGE of DNA
- Mobility Shift Assay
- Methylation & Uracil Interference Assays
- Maxam & Gilbert Sequencing
- Manual Sequencing
- In Gel Enzyme Reactions
- Heteroduplex Analysis
- Gel Preparation for Native PAGE of DNA
- Gel Electrophoresis of PCR Products
- DNase I Footprinting
- DNA/RNA Purification from PAGE Gels
- DNA/RNA Purification from Agarose Gels – Electroelution
- Differential Display
- Denaturing Polyacrylamide Gel Electrophoresis of DNA & RNA
- Conformational Analysis
- Automated Sequencers
- Analysis of DNA/Protein Interactions
- Agarose Gel Electrophoresis of DNA and RNA – Uses and Variations
- Agarose Gel Electrophoresis of DNA and RNA – An Introduction
