Post Electrophoretic Analysis Articles
Manual Sequencing
DNA sequences are determined by a two-step process. In the first step the sample DNA is used, either directly or as a template, to generate sets of fragments. Each set contains multiple lengths of DNA, all of which end in one (or sometimes two) of the four nucleotide bases. These fragments are generally radiolabeled to facilitate detection.
In the second step, the fragments are separated on a denaturing PAGE gel. Each band on the gel represents a position in the DNA sequence, and each position appears only in the fragment set which terminates in the correct base(s).Autoradiography is performed on the gel to visualize the bands. The figure below shows the generation of fragments by two methods, and the gel pattern which results from the electrophoretic separation of these fragments.
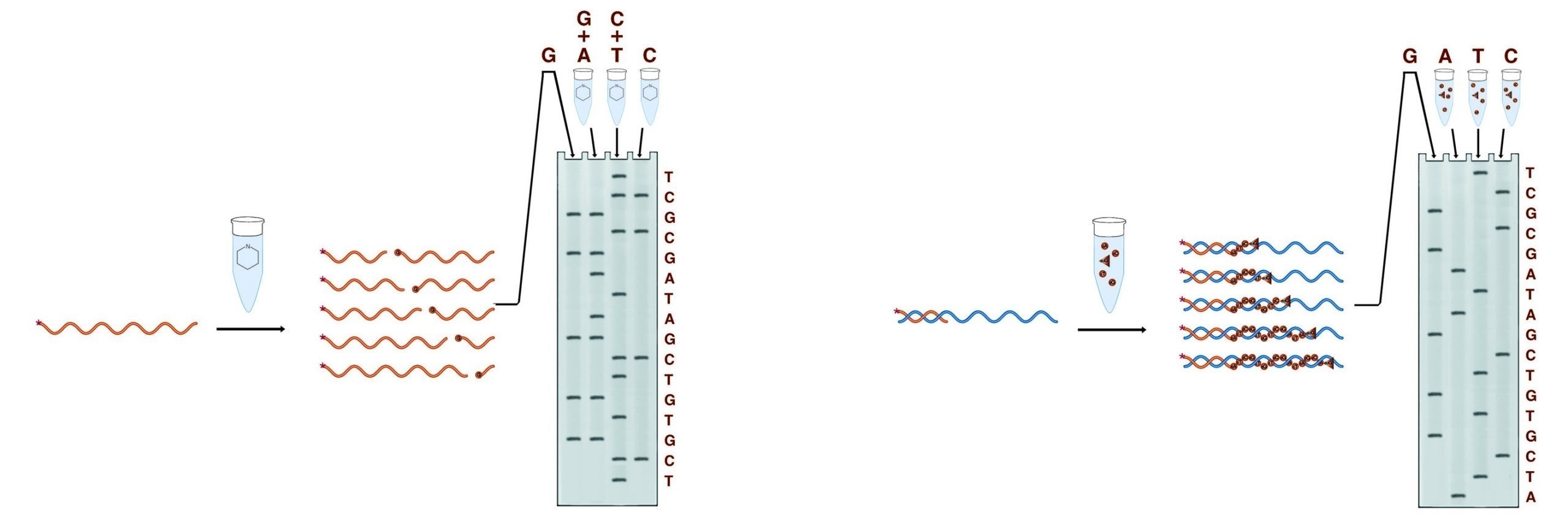
The two methods of conducting sequencing analysis. The Maxam and Gilbert method employs a set of cleavage reactions to generate the necessary fragments while the Sanger method employs a polymerase.
Of the two methods used to generate the fragments for sequencing gel analysis, the Maxam & Gilbert chemical cleavage method was the first widely used. Although no longer predominating, Maxam & Gilbert sequencing possesses significant advantages in certain applications. The four sets of reactions involved in this method cleave the DNA at specific bases or base sets to produce four sets of fragments. No prior knowledge of the DNA sequence is required as the sample DNA is itself processed into fragments. Maxam & Gilbert sequencing is thus useful for sequencing fragments of completely unknown sequences and is essential for footprinting protocols.
Sanger dideoxy terminator sequencing is currently the most widely used chemistry. Although it requires prior knowledge of at least 15 - 20 bases of the sample sequence, it is far less laborious, and more reliable than Maxam & Gilbert sequencing, particularly for long substrate sequences. In this system, the sample DNA is used as a template for a DNA polymerase, typically a bacteriophage polymerase (T4 or T7). Four polymerase reactions are set up for each substrate, each containing enzyme, primer, and sample DNA, along with dNTPs. In addition, each reaction contains one of the four dideoxy NTPs. Dideoxy nucleotides do not have a 3' OH. They are linked to the growing DNA chain through their 5' OH, and the chain stops there for lack of a 3' OH to link to the next base. Each reaction contains one of the four bases as a dideoxy NTP, thus each reaction will contain only fragments which terminate at that base. The proper balance of the levels of DNA, primer, enzyme, ddNTP, and dNTP allows reactions that can give readable sequence out to 1500 bases from the primer. In most dideoxy sequencing, 35S labeled dNTP(s) are included in the polymerase reactions to label the fragments produced. Alternatively, 33P dNTPs or 32P labeled primers can be used.
NEXT TOPIC: Maxam & Gilbert Sequencing
- Using PAGE to Determine Nucleic Acid Molecular Weight
- SSCP Analysis
- Sanger Sequencing
- Sample Preparation for Native PAGE of DNA
- Sample Prep for Denaturing PAGE of DNA
- S1 Mapping
- Run Conditions in Denaturing PAGE
- RNA Mapping
- RNA Electrophoresis
- Ribonuclease Protection
- Restriction Digest Mapping
- Primer Extension
- Preparing Denaturing DNA & RNA Gels
- Preparation of Denaturing Agarose Gels
- Preparation of Agarose Gels
- Pouring Sequencing Gels
- PFGE and FIGE
- PCR Analysis: Yield and Kinetics
- PCR Analysis: An Examination
- Native PAGE of DNA
- Mobility Shift Assay
- Methylation & Uracil Interference Assays
- Maxam & Gilbert Sequencing
- Manual Sequencing
- In Gel Enzyme Reactions
- Heteroduplex Analysis
- Gel Preparation for Native PAGE of DNA
- Gel Electrophoresis of PCR Products
- DNase I Footprinting
- DNA/RNA Purification from PAGE Gels
- DNA/RNA Purification from Agarose Gels – Electroelution
- Differential Display
- Denaturing Polyacrylamide Gel Electrophoresis of DNA & RNA
- Conformational Analysis
- Automated Sequencers
- Analysis of DNA/Protein Interactions
- Agarose Gel Electrophoresis of DNA and RNA – Uses and Variations
- Agarose Gel Electrophoresis of DNA and RNA – An Introduction
