Post Electrophoretic Analysis Articles
Analysis of DNA/Protein Interactions
The binding of proteins to specific DNA sites is an important mechanism of cellular regulation. Numerous techniques have been developed to analyze the interactions of regulatory proteins with DNA. Three techniques are presented here which analyze the site of protein binding on the DNA ("Footprint" Analysis).
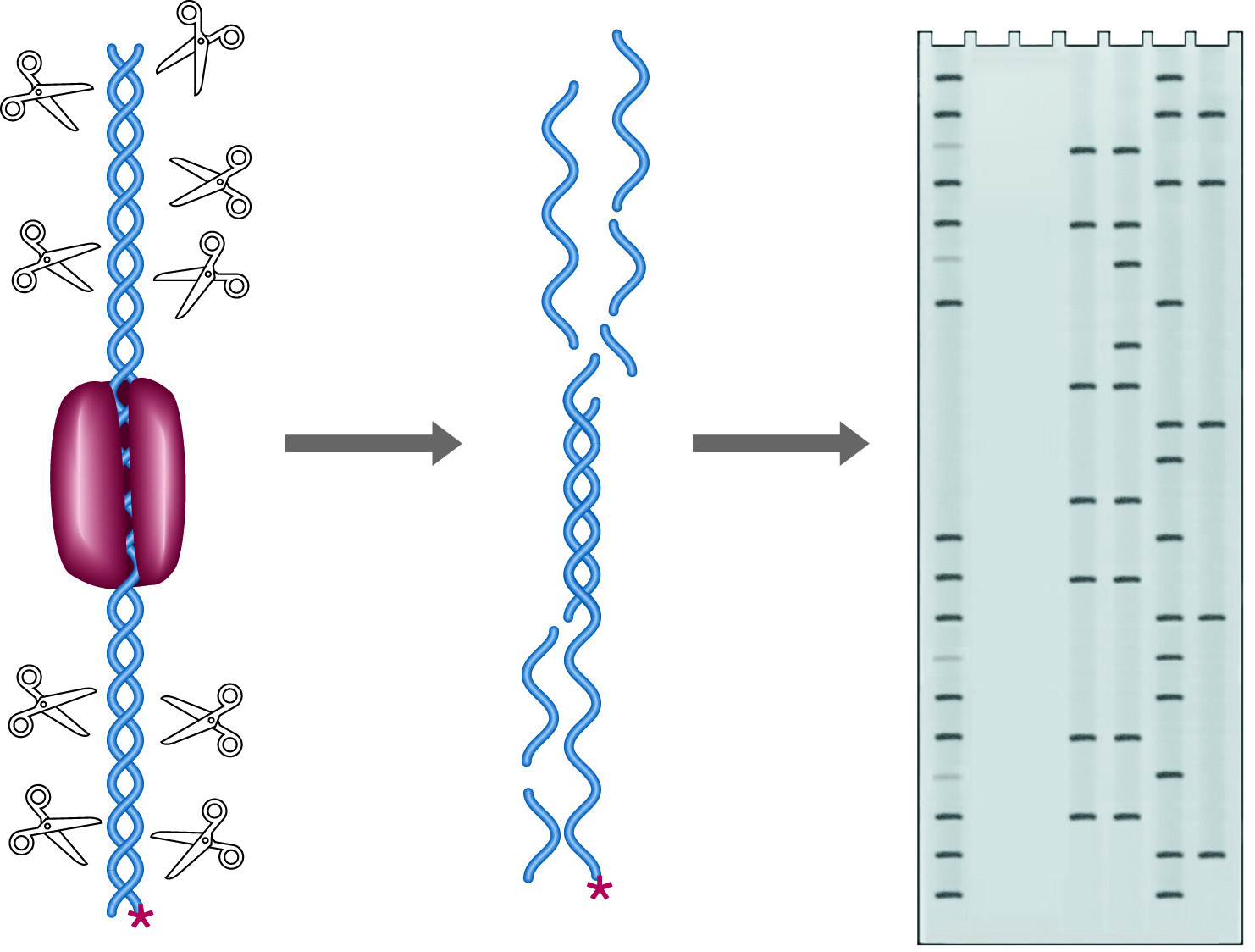
A DNA binding protein will attach itself only to a specific, characteristic sequence on the DNA, and once bound, such a protein occludes the access by other proteins to the binding site. This regional protection is known as a protein's "footprint." The footprint of a particular binding protein may be accurately determined by treating a DNA-protein complex with the enzyme DNase I at low concentrations. DNase I will introduce "nicks" (single strand cuts) in the double stranded DNA, everywhere but in the footprint region. If the DNA is end labeled with 32P, the labeled fragments produced will end at a range of sites which covers the entire starting DNA molecule, excluding the footprint area (See figure below). Separation of these fragments on a denaturing gel yields a ladder of fragments with a gap corresponding to the footprint site. A Maxam & Gilbert sequencing ladder run as a control will allow assignment of the footprint with 1-2 base accuracy.
In addition to mapping the site of the footprint, it is possible to determine which residues within the site are important for binding. This is done using uracil or methylation interference. In these techniques, the nucleotides in the binding site are modified at an average of one site per molecule. This is done either with a methylation reaction which modifies guanine and adenine residues, or by synthesis in the presence of dUTP which substitutes uracil for thymine. The modified fragments are bound to the protein and run on a mobility shift assay which separates bound DNA from unbound DNA. Bound and unbound fractions are separately treated with reagents which cleave the DNA at the modified bases. As with DNase I mapping, end labeled DNA is used, and the resulting fragment lengths reflect the positions of modified bases. Bases which are not found to be modified in the bound fraction are important for protein binding.
NEXT TOPIC: DNase I Footprinting
- Using PAGE to Determine Nucleic Acid Molecular Weight
- SSCP Analysis
- Sanger Sequencing
- Sample Preparation for Native PAGE of DNA
- Sample Prep for Denaturing PAGE of DNA
- S1 Mapping
- Run Conditions in Denaturing PAGE
- RNA Mapping
- RNA Electrophoresis
- Ribonuclease Protection
- Restriction Digest Mapping
- Primer Extension
- Preparing Denaturing DNA & RNA Gels
- Preparation of Denaturing Agarose Gels
- Preparation of Agarose Gels
- Pouring Sequencing Gels
- PFGE and FIGE
- PCR Analysis: Yield and Kinetics
- PCR Analysis: An Examination
- Native PAGE of DNA
- Mobility Shift Assay
- Methylation & Uracil Interference Assays
- Maxam & Gilbert Sequencing
- Manual Sequencing
- In Gel Enzyme Reactions
- Heteroduplex Analysis
- Gel Preparation for Native PAGE of DNA
- Gel Electrophoresis of PCR Products
- DNase I Footprinting
- DNA/RNA Purification from PAGE Gels
- DNA/RNA Purification from Agarose Gels – Electroelution
- Differential Display
- Denaturing Polyacrylamide Gel Electrophoresis of DNA & RNA
- Conformational Analysis
- Automated Sequencers
- Analysis of DNA/Protein Interactions
- Agarose Gel Electrophoresis of DNA and RNA – Uses and Variations
- Agarose Gel Electrophoresis of DNA and RNA – An Introduction
